Genome Sequencing
The genome sequence of Plasmopara viticola has been obtained from isolate INRA-PV221. This isolate was collected in 2009 from a grapevine leaf lesion in a vineyard in the Bordeaux region (Blanquefort, France).
1) First assembly version using short-reads (Dussert et al. 2016)
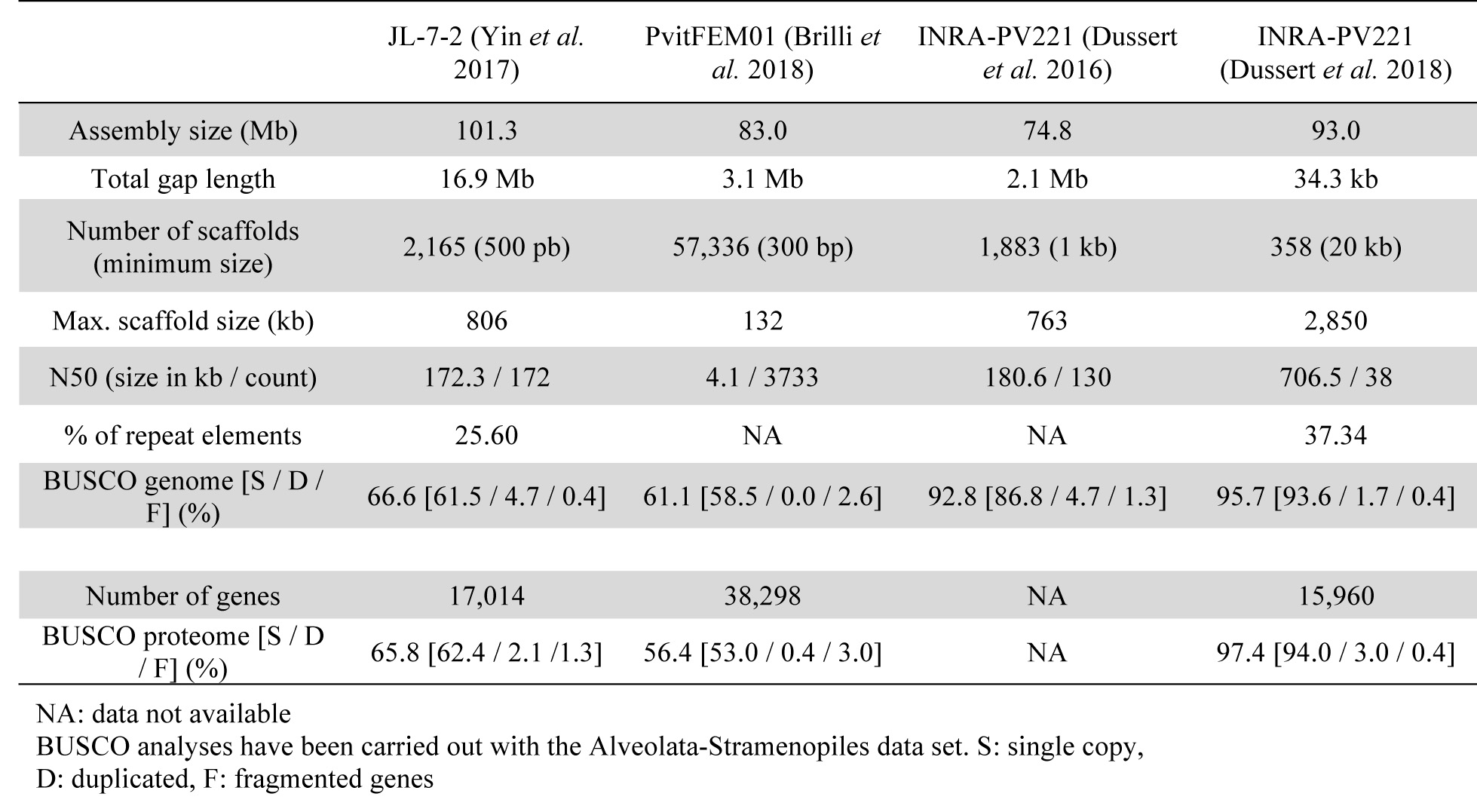
The first draft genome of P. viticola was published by INRA in 2016. A paired-end and two mate-pair libraries (3 and 8-kb inserts) were sequenced. The final assembly included 1,886 scaffolds (maximum size: 763.2 kb), for a total assembly size of 74.8 Mb.
2) Seconde reference genome using long reads at deep coverage (Dussert et al 2019)
We sequenced the INRA-PV221 isolate using PacBio long reads on a RS II sequencer with the P6-C4 chemistry at the GeT-PlaGe facility (Toulouse, France). This sequencing produced 2.6 million reads (mean size: 8,072 bp, total data: 21.2 Gb), corresponding to a mean genome coverage of 185X.
This long-read data allowed us to obtain a new assembly of 92.94 Mb with high continuity (358 scaffolds, N50 of 706.5 kb, max. scaffold size of 2.85 Mb) due a better resolution of repeat regions.
The assembly also showed a high level of gene completeness (95.7% using BUSCO with the Alveolata-Stramenopiles dataset), and notably recovered 1,592 genes coding for secreted proteins and potentially involved in plant-pathogen interactions. Compared to the previous short-read draft, it represented a 70% increase of detected secreted proteins.
Dussert and al, 2019, Genome Biol. Evol. 11(3):954–969, March 7, 2019A High-Quality Grapevine Downy Mildew Genome Assembly Reveals Rapidly Evolving and Lineage-Specific Putative Host Adaptation Genes, doi:10.1093/gbe/evz048
3) Third reference genome using a diploidawre assembly of the P.viticola INRA-Pv221 genome :
Article in submission.